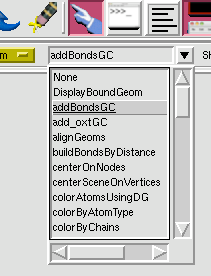
1. Click Picking Commands. It's the hand pointing to the upper left.
2. Choose 'addBondsGC' from the first drop-down menu that appears.
3. Click the unbound atom, a yellow ball should appear upon it along with a label.
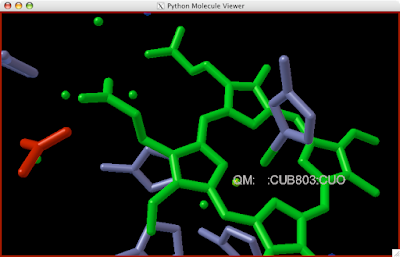
4. Click the atom to which you think it should be bound. A line will be drawn betwixt the atoms.
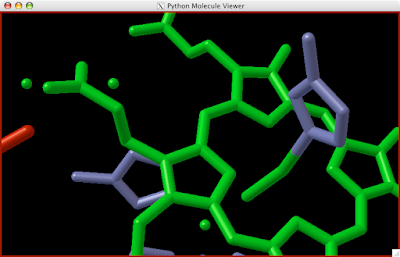
5. Repeat until proper affect achieved.
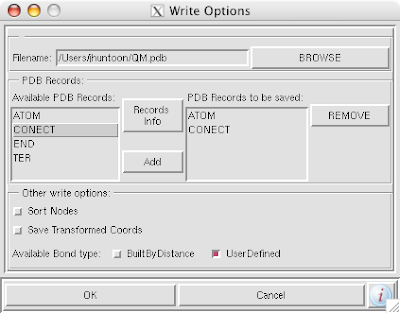
note: if you would like to keep these changes, be sure to save the molecule with the connect records as well as the 'UserDefined' bond types: