Don't try to load that entire biological subunit, a whole virus file could contain a million atoms. You're going to crash your computer or at best move very slowly.
--This is for viruses with icosahedral symmetry similar to poliovirus and rhinovirus. For other icosahedral symmetries, some modifications may need to be applied.--
So here's the best thing to do:
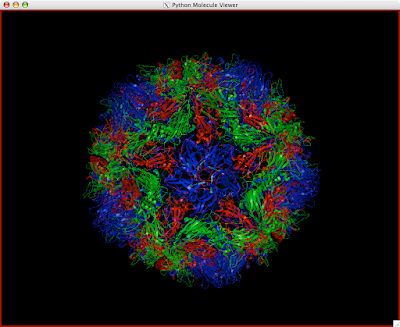
1. Load your desired molecule. I am using the great 2plv, or poliovirus protomer. Poliovirus is a pretty simple structure, as you will see.
2. Open Vision, by clicking the button left of the sunglasses.
3. Load the SymServer Library from Libraries--> Load Libraries--> SymLib
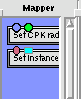
4. From the PMV tab drag down your molecule's node and the 'set instances' node onto the blank canvas.
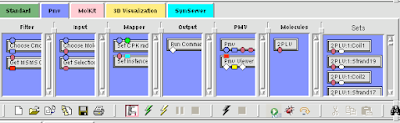
5. From the 'SymServer' tab, drag down the 'Icosahedral 1' node onto the canvas.
your map should look like this now:
6. DoubleClick the 'Icosahdral 1' node. Connect the 'input Ports' node to the '5-fold' node (blue box to blue box).
7. Connect your molecule's node to the 'Set Instances' node (purple oval to purple oval) and connect the 'Icosahedral 1' node to 'Set Instances' node as well ( blue box to blue box)
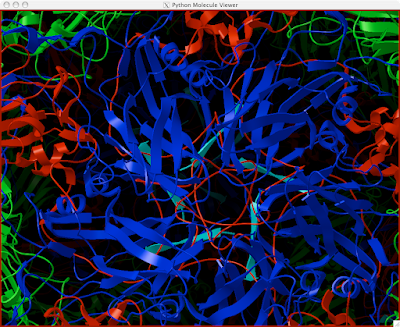
You should be golden now! You can make surfaces and color or anything you want, and the great thing about it is that your really only doing this to molecule instead of 60! This way, hardly any memory is used up!
-Jon Huntoon