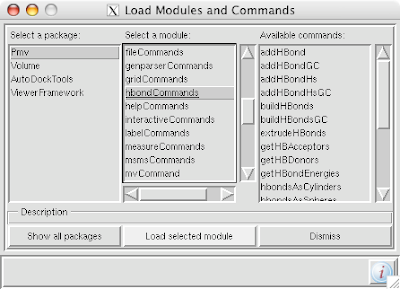
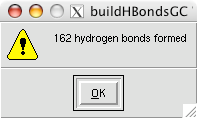
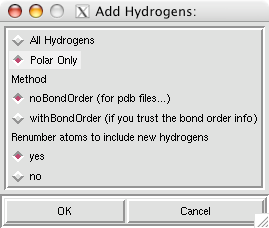
Here is a real quick trick: if you want the lighting to instantly be bright on your object (by eliminating the depth cue), hit the "D" key and voilà.
1. Press "D" key.

Tips, Tricks, and Tutorials for Python Molecular Viewer


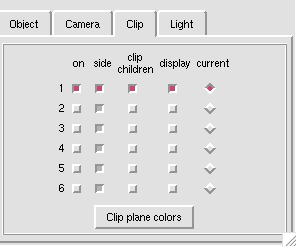
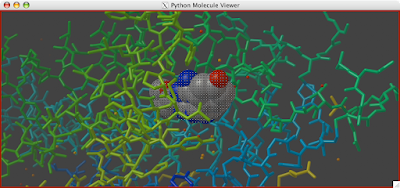
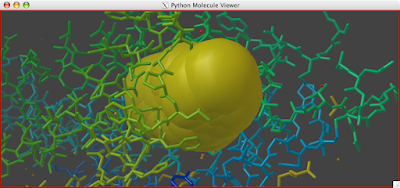
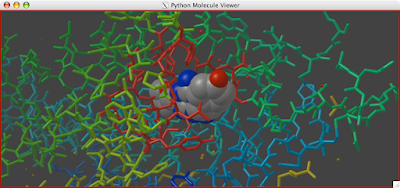
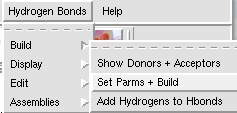
 A clipping plane is an excellent way to explore the interior of a protein structure, while also giving reference to the exterior. Sometimes, you can even find complex tunnels that bore into the interior of a protein. To clip in PMV:
A clipping plane is an excellent way to explore the interior of a protein structure, while also giving reference to the exterior. Sometimes, you can even find complex tunnels that bore into the interior of a protein. To clip in PMV: