How you decide to distinguish between chains in a molecule is really up to taste, but I think there is room for improvement in legibility. Most scientists distinguish chains by color. I do too. But we can go farther, and perhaps, do better.
By default, molecular surfaces are calculated with disregard for individual chains:
This is actually more true to 'real life' or 'in situ'. The water molecules defining the surface of a protein have no regard for chains. They simply pass over. But we operate 'in silico', and afforded the opportunity to distinguish with clarity between the molecular surfaces of chains. Perhaps for the better in the end for discovery- or at least another arrow in our quiver.
Here is a side by side comparison:
I think it is clear from the screen shots above that calculating the molecular surface per chain can be a positive augmentation of the representation.
So, without further adieu, I present the simple trick:
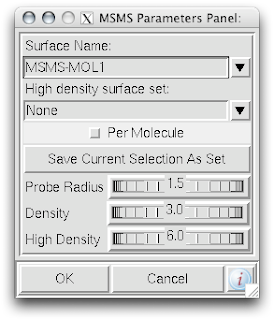 1. While in the computeMSMS popup menu, de-select the 'Per Molecule' check box.
1. While in the computeMSMS popup menu, de-select the 'Per Molecule' check box.2. In that same menu, be sure to rename the 'Surface Name' for each chain.
That's it!
Another quick tip is that you can reach the ComputeMSMS menu by right-clicking the radio buttons in dashboard:
Enjoy!
JON HUNTOON





















