1 Open DejaVu.
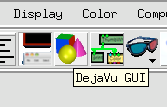
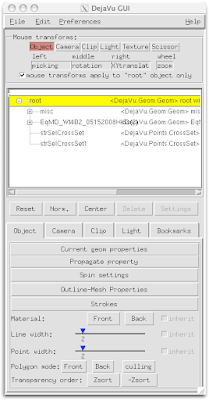
2 Select Spin Settings.
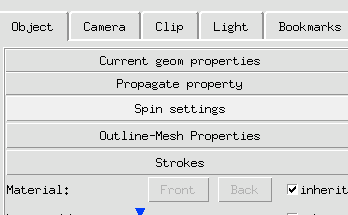
3 Click Spin button to engage spinning.
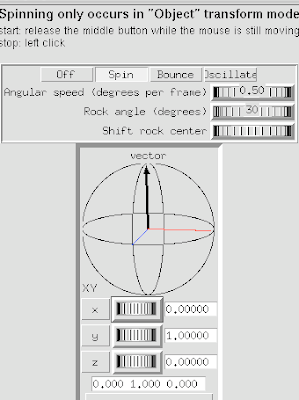 4 Adjust using the Vector axes arrows.
4 Adjust using the Vector axes arrows. 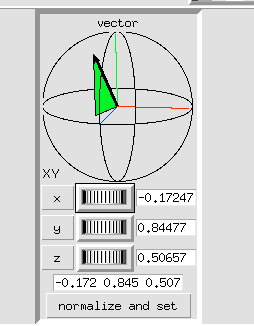
Tips, Tricks, and Tutorials for Python Molecular Viewer
| All you need is a PDB and an idea and we'll build an actual three-dimensional physical structure you can hold in your hand and pass from person to person. Actually we can use almost any file format to create your molecular model. Also, you may build your own model in PMV or pymol and send the session file our way. The cost of these models is determined by the volume of the model, the amount of Superglue used to hold it together, and the time it took to make. That is, $5/cubic inch, $10/ounce of superglue, and $25/ hour. Contact Jon Huntoon by phone (858) 784-2751 or by email huntoon@scripps.edu. Everything we make is custom. A normal model will take somewhere between 1 and 10 hours of modeling time, 1 to 50 cubic inches of material and 0.5 to 5 ounces of superglue, putting the price range at $50 to $500 most of the time. Please note there will be a 89.5% overhead fee for all models made for researchers outside of The Scripps Research Institute. |

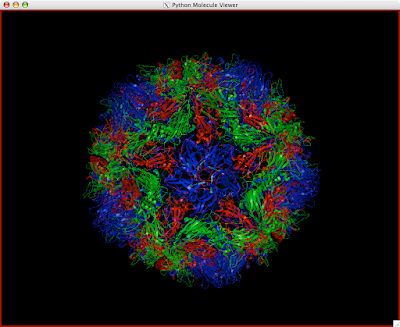
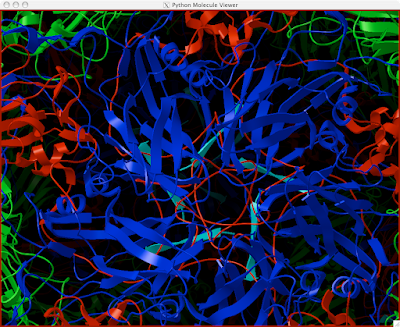

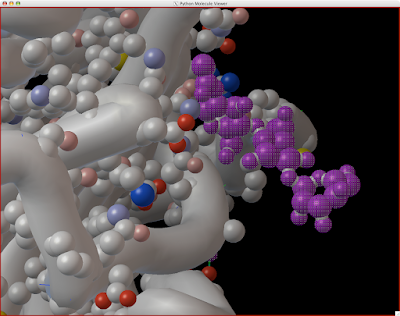
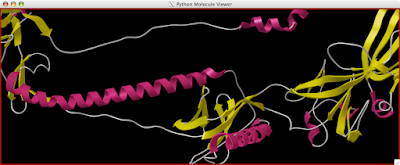


Here is how you can read and display an electron density map. These are important to look at if you are to be critical of a particular PDB structure.
1a Load PDB

1b Download the corresponding CCP4 file from PDB.
2a Create a Vision Network.
 (middle button, boxes & lines, not the sunglasses button)
(middle button, boxes & lines, not the sunglasses button)
2b Load the Vis and Vol Libraries from Libraries--> Load Libraries....
2c Drag the following 'nodes' on to the blank 'canvas':
- 'Read Any Map' (Load CCP4 here) -from Vol Library (grey)
- 'Sample' (use a level 3 or two) - from Vol Library
- 'UT iso-contour' From Vol Library
- 'Ind Polygons' -from Vis Library (yellow)
- 'PMV Viewer' - from PMV Library (purple-ish/blue?... hmm. Periwinkle!)
(it helps to use the search box  to find these nodes)
to find these nodes)
Connect these nodes by dragging lines from the output ports (bottom square things) to the input ports(top square things) (colors should correspond [mostly]):
3 Double-click the UT iso-contour 'node' to see the isocontour graphs. Looks like this:
4 Drag the black vertical line to Pick an iso-value that suits your purposes. It will be hairy, so you may want to...
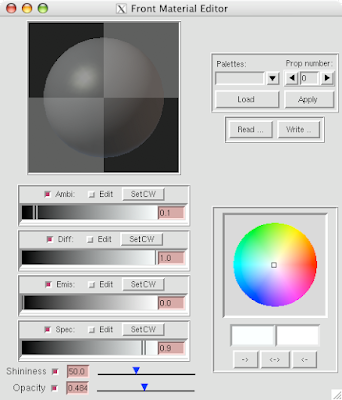
5 Open DejaVu  (it's the left-erly button with a cube, sphere and pyramid) to adjust opacity and or clipping of the IndPolygons geometry you just created!
(it's the left-erly button with a cube, sphere and pyramid) to adjust opacity and or clipping of the IndPolygons geometry you just created!
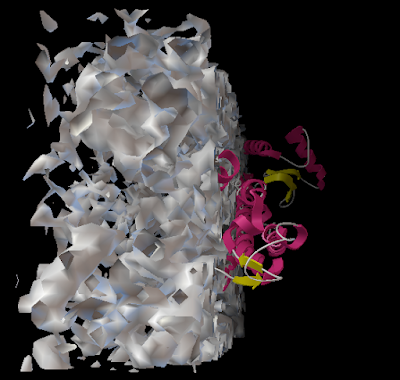
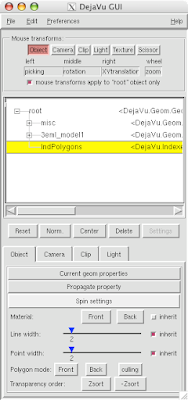
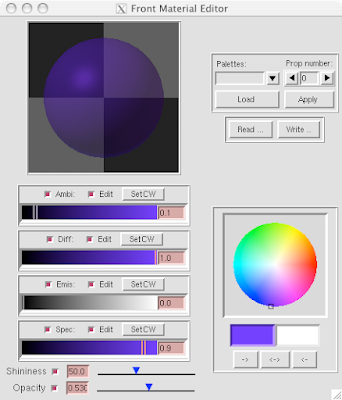
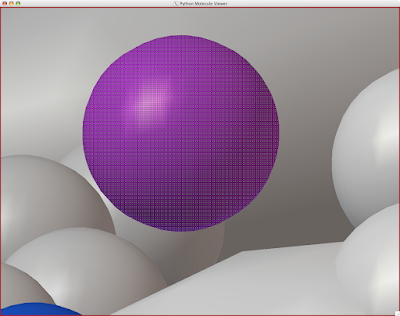
And you can end up with something like this or (probably) better. Explore around. Have fun. Get good.
--
Jon Huntoon