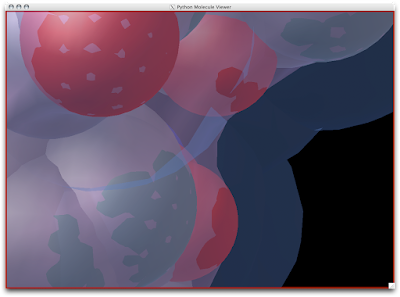
David Goodsell as you may know, is an exceptional artist and brilliant communicator. His watercolors have been published in magazines, journals, and books (The Machinery of Life
Lucky us, PMV has an auto 'David Goodsell' feature.
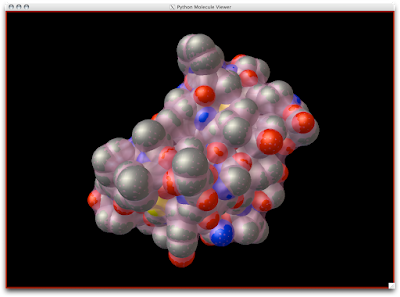
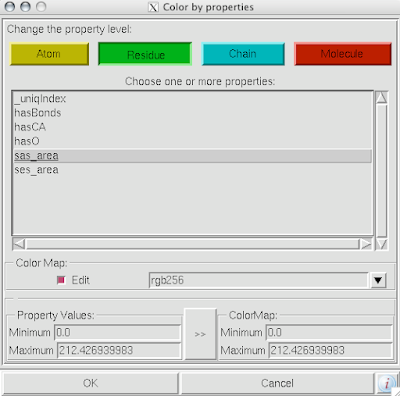
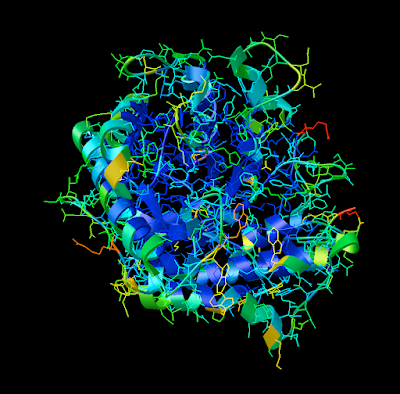
1. Firstly, there is the "DG Colors" under the color menu. This coloring is similar to atomic but the intensity of the red and blue on oxygens and nitrogen respectively corresponds to the charge of the residue upon which the atom resides. For example, a highly negatively charge oxygens on glutamate will be deep red, where as the less negatively charged oxygens on threonine will be pink. Conversely, the strongly positive charged nitrogen upon the lysine will be dark blue while the less positive nitrogens on histidine are sky blue. A clever coloring scheme that incorporate charge and atom property is one way in which the David Goodsell style conveys so much information without becoming too busy.
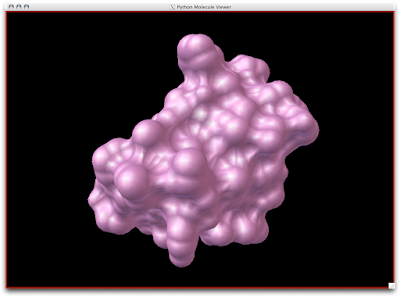
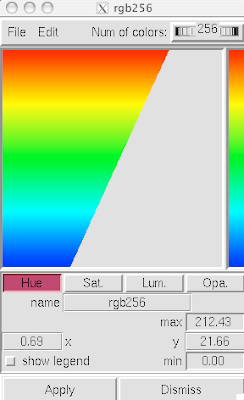
2. Now that your models are colored according the custom DG scheme, it is time to emulate the subtle beauty of his water colors. Turn on the 'cartoon mode'.
adjust if you wish:
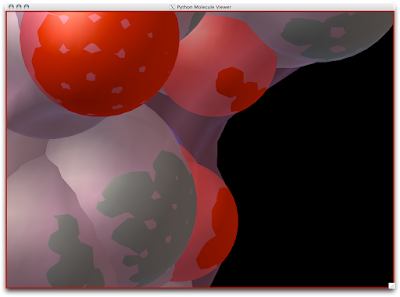
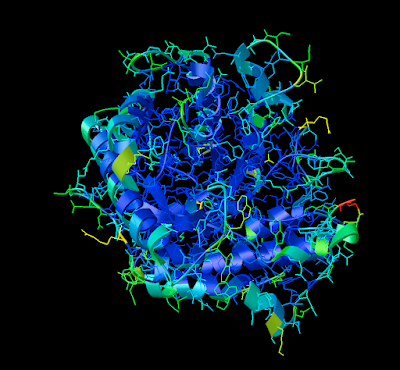
3. Now you are officially David Goodsell-esque. The last recommendation I have is to increase the anti-aliasing to at least 4. Beyond 8, the anti-aliasing tends to suck too much CPU power.
ENJOY!